Friday, December 11, 2015
Trying new blog software
http://richardsprague.com
I'm trying to rebuild a personal site using my own software and it's going more slowly than I'd like, so please be patient. One of these days it'll be great.
Meanwhile, if you are here hoping to find more info about my work with the microbiome, please contact me directly. I'm working on a major new project to make it easy for normal people to analyze and understand their microbiome test results, and I would be grateful if you could send me your data in return for some cool analysis. Just email me.
Friday, October 16, 2015
Hunter-gatherers sleep like me
Anthropologists Gandhi Yetish, Hillard Kaplan and their colleagues just published the results of some experiments that show hunter gathers get much less sleep than the eight hours we supposedly need. In fact, their sleep patterns closely resemble mine, despite the conventional wisdom that my average 6.5 hours per night is too little. (There’s a nice summary by Anahad O'Connor in a New York Times Blog post)
A long-time fan of Zeo, I have carefully measured my sleep for many years and I know that my lifetime average is pretty close to 6.5 hours. That’s real sleep, measured by a brain wave detector strapped to my head. Like other lazy people, I sometimes lay in bed longer than that, but it’s a rare occasion when my sleep duration is longer than 7 hours, even when I’m loaded with potato starch to grow serotonin-helping bifidobacterium.
Because the new study appears to contradict the rest of conventional scientific wisdom about the importance of 8+ hours of sleep, I read it carefully, along with the data collected to see if I could spot any problems. So far I think everything adds up:
Plenty of participants: 100 people, spread among male/female at different ages, including some fairly old: 60+
Three separate, unrelated societies: from both Africa and South America, it’s hard to argue these people are somehow related at anything other than being hunter-gatherers.
Week-long observations: You might want a study like this to go on for weeks or years, but I think the duration, from a week to a month per person was just fine.
Good self-tracking hardware: the anthropologists used the Philips Actiwatch 2, strapped to subjects’ wrists with a tamper-proof hospital band. These are well-studied, medical-grade wearables and although they use actigraphy information, not perfect because it’s based on movements in the night, if anything these devices tend to overestimate the amount of sleep. I skimmed the data from the study and it looks good.
The authors conclude that ambient temperature, not daylight, is the most important signal that tells these hunter-gatherers it’s time to sleep. They note that these people sleep no the ground on skin mats, inside huts or in the outdoors, often covered with lightweight cotton blankets. This isn’t all that different from camping, when I tend if anything to sleep more.
Interestingly, when Zeo studied 5000 of their users back in 2011, they found an average sleep time of something closer to 8 hours, with my 6.5 hours falling out of the 95% confidence interval, making me (and the hunter gatherers) real outliers.
Note that although I rarely sleep longer than 6.5 hours, I feel great in the morning and I’m generally alert and feel reasonably fresh all day. Like the hunter-gatherers, I don’t nap and I rarely suffer from insomnia.
I’ll be watching the follow-ups to this research carefully but for now I’ll be much more satisfied that my current level of sleep is just fine.
Monday, September 28, 2015
Microbes on my skin
So today, imagine my surprise when they sent me a nice note announcing that they’d re-run my latest skin sample and found something! The web site version only found two phyla, which at first didn’t seem very interesting:
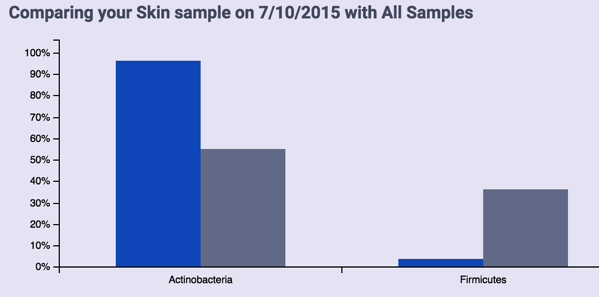
The sample shows up as only two taxa: Actinobacteria (96%) and Firmicutes (the rest). Fortunately, by downloading the taxonomy data and converting to Excel, I was able to get more details. Here’s my skin at the genus level:
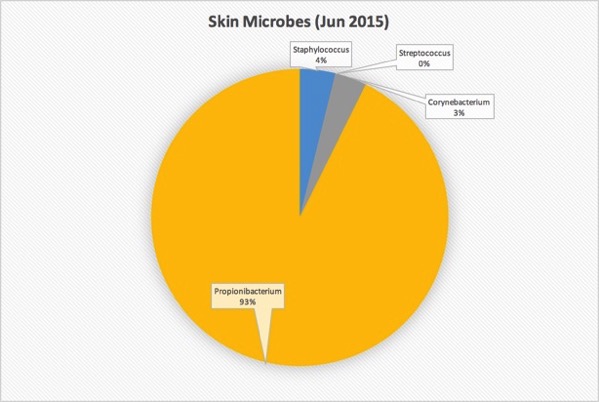
The vast, vast majority of microbes behind my ear are Propionibacteria, which appears to be true for most people and most skin types. This genus contains the special P. Acnes species known to be linked to acne. The species level information in my case only identified about half of all the taxa found in my sample, so although none of what was found was P. Acnes, it’s possible that it’s lurking in there someplace.
Since learning more about the microbiome, I’ve stopped using hand sanitizers, switched to shampoo that is free of sodium laurel sulfate, and I take other precautions to ensure i have the most “natural” skin microbiome I can get, but I don’t know if those actions have helped (or hurt). The only way to find out will be to send another sample so I can compare. Now that I know uBiome is able to process my skin type, I’ll be sure to do that.
Tuesday, September 15, 2015
I'm a secretor
The Gut Guardians podcast interview with Alanna Collen included an interesting reference to the FUT2 gene, which the podcast hosts says has been linked to response to high fiber diets. One of the alleles, referred to as the non-secretor type, offers a genetic immunity to infection by the Norwalk Norovirus, also known as the “cruise ship virus”.
Alas, I’m not immune to that particular virus, but if you have to choose I think it’s better to have the “secretor allele”, like me. Overall, secretors seem less susceptible to many influenza strains and pathogenic bacteria, perhaps due to our better response to high fiber.
What’s especially interesting is that how FUT2 also seems to be associated with your microbiome. Secretors have noticeably different levels of various bacteria, including missing Bifido species that are known to play a role in health. This is another example of how genes don’t have the final say: you may not get the full benefits of being a secretor if you don’t eat enough fiber.
If you have your 23andme results, you can check your FUT2 status too.
Tuesday, August 25, 2015
Measuring my uBiome diversity
While there are few agreed-upon standards for what constitutes a good or bad microbiome, nearly everyone agrees that a diverse microbiome is better than one that is less diverse. How can we measure diversity?
Ecologists have been interested in this question for a long time and they’ve developed several metrics to describe how diverse a particular ecosystem is compared to another. This is my very tentative first step to try to adopt those metrics to my Ubiome results.
You could simply count all the different species (or genera or phyla) in a sample and track that over time. The more unique species, the more diversity. Here’s what that looks like for me:
In this case, all I did was plot the raw number of taxa that uBiome found at each tax_rank. That’s about 60 species in my latest sample. But since 16S technology doesn’t capture all the species information, or the genera or phyla information for that matter, a simple count of the number of organisms detected is not terribly useful. In my case, uBiome identified between 90-97% of all the phyla in my samples, but only between 49-51% of the species. That makes apples-to-apples comparisons difficult to interpret.
Ecologists have suffered from this problem for a long time, and they came up with a few metrics to get around the it. They start by considering what it means to say something is more diverse than another. Consider a forest that has 1,000 trees in it. If all 1,000 trees are, say, aspen trees, then that forest is not as diverse as another one that might also have 1,000 individual trees but, say, 1,000 unique species.
There’s a unit of information called the Shannon number, after the information theorist Claude Shannon, who was the first mathematician to systematically try to measure information. To Shannon, whose work was concerned with code breaking in World War II, a radio signal that carries information (i.e. a code) will be slightly different from one that is random noise. He applied a specific formula to tell how different a signal looks compared to random noise, a variation of which can be applied to an ecosystem to tell how different it is from one that is completely dead (0) or has nothing but the same or similar organisms.
I use a slightly more ecologically interesting version of the Shannon number, called the Inverse Simpson number, that looks at the total number of unique life forms in an ecosystem and then weights each by the number of individuals of that type of species. Conveniently, these functions are all available in the R “Vegan” package. Here’s how I set up my R environment to do the calculations:
allSprague <- read.csv("spragueResultsThruJun2015.csv")
allGenus <- allSprague[allSprague$tax_rank=="genus",]
allPhylum<-allSprague[allSprague$tax_rank=="phylum",]
allSpecies<-allSprague[allSprague$tax_rank=="species",]
allSamples <- allGenus[,-(1:2)]
require("vegan",quietly=TRUE)dV <- sapply(allSamples,fisher.alpha)
names(dV)<-c(as.Date("2014-5-23"),as.Date("2014-6-10"),as.Date("2014-10-23"),as.Date("2015-1-15”),as.Date("2015-2-20"),as.Date("2015-4-20"),as.Date("2015-4-27"),as.Date("2015-6-15"))
g<-names(allSprague)
dPDates<-as.Date(names(dV))Here is my diversity as an Inverse Shannon number:
#genus diversity
dG <- sapply(allGenus[,-(1:2)],diversity,index="invsimpson")
plot(dG~dPDates,main="Genus Diversity",xlab="",ylab="Inv Simpson Diversity",xaxt="n")
axis(1,at=dPDates,labels=g[-(1:2)],las=2)
abline(lm(dG~dPDates),col="red")
What does it mean? Apparently, my overall Genus diversity has declined in the past year, though it bumps up and down so much that it’s hard to see a real pattern with so few data points. It’ll be interesting to compare my diversity to a few other people using this function.
My apologies for the super-technical nature of this post, especially with the un-cleaned R source code. I’m just throwing it on the blog so I discuss it with people who are way more knowledgable than I am and can hopefully guide me to something better. I’ll have much more to say later, after I actually understand what I’m talking about.